This class implements a rigid-tendon muscle-torque-generator (MTG) for a growing list of joints and torque-directions. For a detailed description of the MTGs available and the automatic fitting routine (implemented in TorqueMuscleFittingToolkit) please see the publication: M.Millard, A.L.Kleesattel, M.Harant, & K.Mombaur. A reduced muscle model and planar musculoskeletal model fit for the synthesis of whole body movements. Journal of Biomechanics. (under review as of August 2018) More...
#include <Millard2016TorqueMuscle.h>
Private Member Functions | |
| void | updateTorqueMuscleCurves () |
| double | calcJointAngle (double fiberAngle) const |
| double | calcFiberAngle (double jointAngle) const |
| double | calcFiberAngularVelocity (double jointAngularVelocity) const |
| double | calcJointAngularVelocity (double fiberAngularVelocity) const |
| void | calcTorqueMuscleDataFeatures (RigidBodyDynamics::Math::VectorNd const &jointTorque, RigidBodyDynamics::Math::VectorNd const &jointAngle, RigidBodyDynamics::Math::VectorNd const &jointAangularVelocity, double activeTorqueAngleBlendingVariable, double passiveTorqueAngleBlendingVariable, double torqueVelocityBlendingVariable, double activeTorqueAngleAngleScaling, double activeTorqueAngleAtOneNormTorque, double passiveTorqueAngleCurveOffset, double maxAngularVelocity, double maxActiveIsometricTorque, TorqueMuscleDataFeatures &tmf) const |
| double | calcFiberAngleGivenNormalizedPassiveTorque (double normPassiveFiberTorque, double blendingVariable, double passiveTorqueAngleCurveOffset) const |
| double | calcBlendedCurveDerivative (double curveArgument, double blendingVariable, double maximumBlendingValue, unsigned int derivativeOrderArgument, unsigned int derivativeOrderBlendingVariable, RigidBodyDynamics::Addons::Geometry::SmoothSegmentedFunction const &curve) const |
| double | calcInverseBlendedCurveValue (double blendedCurveValue, double argGuess, double blendingVariable, double maximumBlendingValue, RigidBodyDynamics::Addons::Geometry::SmoothSegmentedFunction const &curve) const |
| void | updTorqueMuscleSummary (double activation, double jointAngle, double jointAngularVelocity, double activeTorqueAngleBlendingVariable, double passiveTorqueAngleBlendingVariable, double activeTorqueAngularVelocityBlendingVariable, double activeTorqueAngleAngleScaling, double activeTorqueAngleAtOneNormTorque, double passiveTorqueAngleCurveOffset, double maxAngularVelocity, double maxActiveIsometricTorque, TorqueMuscleSummary &updTms) const |
| void | updTorqueMuscleSummaryCurveValues (double fiberAngle, double normFiberAngularVelocity, double activeTorqueAngleBlendingVariable, double passiveTorqueAngleBlendingVariable, double activeTorqueAngularVelocityBlendingVariable, double activeTorqueAngleAngleScaling, double activeTorqueAngleAtOneNormTorque, double passiveTorqueAngleCurveOffset, TorqueMuscleSummary &updTms) const |
| void | updTorqueMuscleInfo (double activation, double jointAngle, double jointAngularVelocity, double activeTorqueAngleBlendingVariable, double passiveTorqueAngleBlendingVariable, double activeTorqueAngularVelocityBlendingVariable, double activeTorqueAngleAngleScaling, double activeTorqueAngleAtOneNormTorque, double passiveTorqueAngleCurveOffset, double maxAngularVelocity, double maxActiveIsometricTorque, TorqueMuscleInfo &updTmi) const |
| void | updInvertTorqueMuscleSummary (double jointTorque, double jointAngle, double jointAngularVelocity, double activeTorqueAngleBlendingVariable, double passiveTorqueAngleBlendingVariable, double activeTorqueAngularVelocityBlendingVariable, double activeTorqueAngleAngleScaling, double activeTorqueAngleAtOneNormTorque, double passiveTorqueAngleCurveOffset, double maxAngularVelocity, double maxActiveIsometricTorque, TorqueMuscleSummary &updTms) const |
Static Private Attributes | |
| static const double | mTaLambdaMax |
| static const double | mTpLambdaMax |
| static double const | Anderson2007Table3Mean [36][14] |
| static double const | Anderson2007Table3Std [36][14] |
| static double const | GymnastWholeBody [24][12] |
Friends | |
| class | TorqueMuscleFittingToolkit |
| class | FitTorqueMuscleParameters |
Detailed Description
This class implements a rigid-tendon muscle-torque-generator (MTG) for a growing list of joints and torque-directions. For a detailed description of the MTGs available and the automatic fitting routine (implemented in TorqueMuscleFittingToolkit) please see the publication: M.Millard, A.L.Kleesattel, M.Harant, & K.Mombaur. A reduced muscle model and planar musculoskeletal model fit for the synthesis of whole body movements. Journal of Biomechanics. (under review as of August 2018)
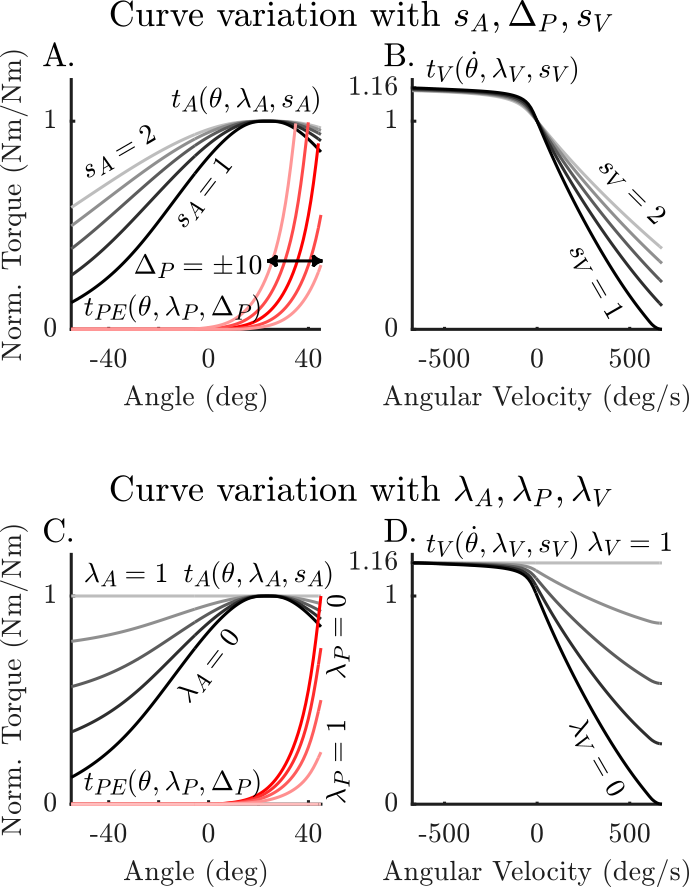
This rigid-tendon torque muscle model provides modeling support for 3 phenomena
- torque-angle curve (
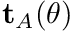 ): the variation of active isometric torque in one direction as a function of joint angle
): the variation of active isometric torque in one direction as a function of joint angle - torque-velocity curve (
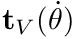 ): the variation of torque as a function of angular velocity
): the variation of torque as a function of angular velocity - passive-torque-angle curve (
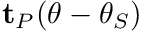 ): the variation of passive torque as a function of joint angle. Here
): the variation of passive torque as a function of joint angle. Here 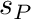 and
and 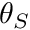 are user-defined scaling and shift parameters.
are user-defined scaling and shift parameters.
each of which are represented as smooth normalized curves that vary between 0 and 1. A series of scaling ( 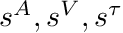 ), shifting (
), shifting ( 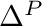 ), and blending (
), and blending ( 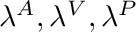 ) variables have been introduced to these curves to make it possible to easily fit these curves to a specific subject. These fitting variables can be set/fitted by making use of the functions in this class. Alternatively these fitting variables can be automatically adjusted (using IPOPT) by making use of the TorqueMuscleFittingToolkit.
) variables have been introduced to these curves to make it possible to easily fit these curves to a specific subject. These fitting variables can be set/fitted by making use of the functions in this class. Alternatively these fitting variables can be automatically adjusted (using IPOPT) by making use of the TorqueMuscleFittingToolkit.
These three curves are used to compute the torque developed  given the angle of the joint
given the angle of the joint  , the angular-velocity of the joint
, the angular-velocity of the joint  , and the activation of the muscle
, and the activation of the muscle 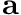 (a 0-1 quantity that defines how much the muscle is turned-on, or activated), and the maximum-isometric torque
(a 0-1 quantity that defines how much the muscle is turned-on, or activated), and the maximum-isometric torque 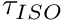
![\[ \tau (\mathbf{a}, \theta,\dot{\theta}) = \tau_{ISO} ( \mathbf{a} \, \mathbf{t}_A(\theta) \mathbf{t}_V(\dot{\theta}/\dot{\theta}_{MAX}) +\mathbf{t}_P(1- \beta (\dot{\theta}/\dot{\theta}_{MAX})) \, ) \]](../../form_180.png)
The damping term 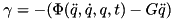 is necessary to supress vibration that will occur as the passive element
is necessary to supress vibration that will occur as the passive element 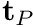 is streched, its stiffness increases, and the natural frequency of the overall system rises. By default
is streched, its stiffness increases, and the natural frequency of the overall system rises. By default 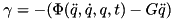 is set to 0.1 which has proven effective for supressing vibration in the trunk segments during a stoop lift in which the stiffness of the lumbar back muscles grows appreciably. This model does not yet provide support for the following phenomena but will in the future.
is set to 0.1 which has proven effective for supressing vibration in the trunk segments during a stoop lift in which the stiffness of the lumbar back muscles grows appreciably. This model does not yet provide support for the following phenomena but will in the future.
- activation dynamics: currently is left to the user
- tendon-elasticity
- muscle short-range-stiffness
All of these characteristic curves are represented using 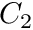 continuous
continuous 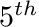 order Bezier curves that have been fitted to the data from data in the literature. In many cases these curves have been carefully edited so that they fit the curves of the original papers, but have more desireable numerical properties for optimal control work. The characterisic curves provided by this class have been fitted to a growing list of data sets: -Anderson Data Set: from Anderson et al. 2007 -Whole-body Gymnast Data Set: from Jackson, Kentel et al., Anderson et al., Dolan et al. and Raschke et al.
order Bezier curves that have been fitted to the data from data in the literature. In many cases these curves have been carefully edited so that they fit the curves of the original papers, but have more desireable numerical properties for optimal control work. The characterisic curves provided by this class have been fitted to a growing list of data sets: -Anderson Data Set: from Anderson et al. 2007 -Whole-body Gymnast Data Set: from Jackson, Kentel et al., Anderson et al., Dolan et al. and Raschke et al.
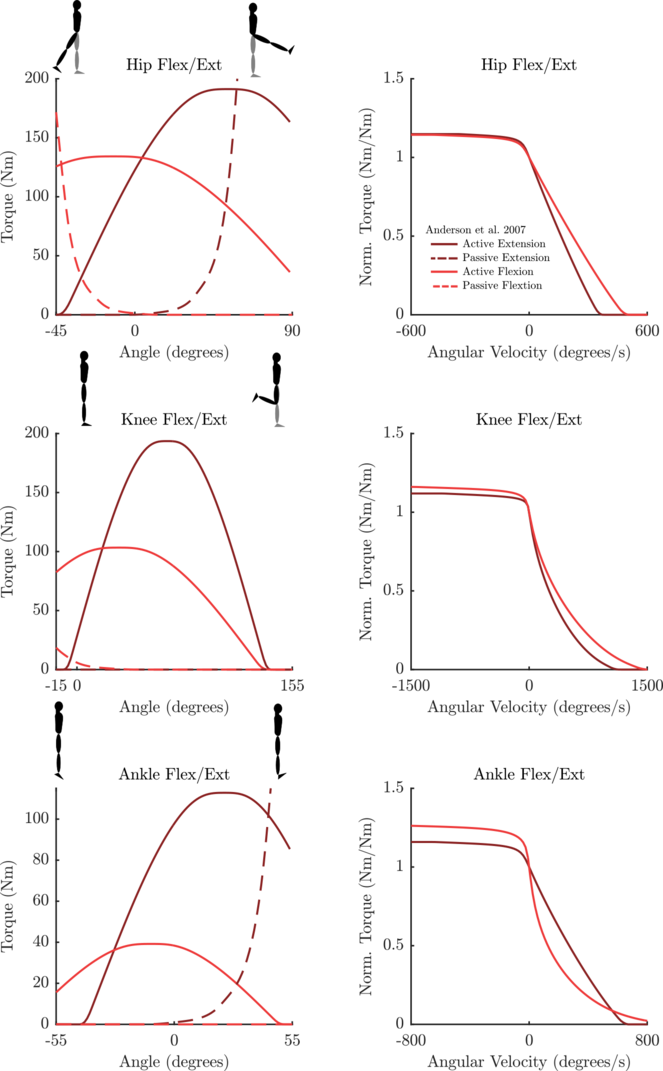
Data Set: Anderson2007
This data set uses the mean value of the coefficients published in Anderson et al. The standard deviation table has also been entered. However, since it is unclear how to use the standard deviation in a consistent way across all joints/parameters this table is not yet accessible through the constructor. This data set includes coefficients for the following
- Number of subjects: 34
- Gender: male and female
- Age: young (18-25, 14 subjects), middle-aged (55-65, 14 subjects), senior (> 65, 6 subjects)
- Joint: hip/knee/ankle
- Direction: extension/flexion
Notes
- Angles are plotted using units of degrees for readability. The actual curves are described in units of radians
- See Anderson et al. for further details.
Data Set: Gymnast
This data set is an attempt at making enough torque muscles for a whole body. Since no single source in the literature comes close to measuring the characteristics of all of the joints, data from Jackson et al., Kentel et al., Anderson et al., Dolan et al, and Raschke et al. have been combined. Since the subjects used in these various studies are wildly different (Jackson et al. measured an elite male gymnast; Kentel et al. measured an elite tennis player; Anderson et al. measured, in the category of young male, a selection of active undergraduate students, Dolan et al from 126 women and 23 men, and Raschke from 5 male subjects) scaling has been used to make the strength of the subject consistent. Scaling coefficients for the lower body, shoulders and elbow, and forearm/wrist have been calculated using measurements that overlapped between datasets. Presently this data set includes curves for 22 torque muscles.
- Number of subjects: 1 elite gymnast (69.6 kg, 1.732 m)
- Gender: male
- Age: 21 years old
- Joint and Directions available
- Ankle: flexion/extension (scaled from Anderson)
- Knee: flexion/extension (from Jackson)
- Hip: flexion/extension (from Jackson)
- Lumbar: active extension curves (
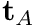 and
and 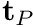 ) from Raschke et al. passive extension from Dolan et al. The torque velocity curve has since been updated using an estimate from the archiecture of the lumbar extensors
) from Raschke et al. passive extension from Dolan et al. The torque velocity curve has since been updated using an estimate from the archiecture of the lumbar extensors - Lumbar: active flexion
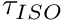 from Beimborn et al.
from Beimborn et al.
- Shoulder: flexion/extension (from Jackson)
- Shoulder: horizontal adduction/abduction (from Kentel, scaled to Jackson's subject)
- Shoulder: internal rotation/external rotation (from Kentel, scaled to Jackson's subject)
- Elbow: flexion/extension (from Kentel, scaled to Jackson's subject)
- Wrist: pronation/supination (from Kentel, scaled to Jackson's subject)
- Wrist: extension/flextion (from Jackson)
- Wrist: ulnar/radial deviation (from Kentel, scaled to Jackson's subject)
- Missing Joint and directions
- Ankle inversion/eversion
- Hip adduction/abduction
- Hip internal rotation/external rotation
- Lumbar extension/flexion
- Lumbar bending
- Lumbar twisting
- Shoulder Adduction
- Shoulder Abduction
- Scapular elevation/depression
- Scapular adduction/abduction
- Scapular upward/downward rotation
In all cases the curves have been fitted to Bezier curves that are constructed using functions in TorqueMuscleFunctionFactory.
Notes
- Angles are plotted using units of degrees for readability. The actual curves are described in units of radians
- Hip and Knee characteristics taken from Jackson. Ankle extension is from Anderson et al., scaled using Jackson-to-Anderson hip/knee strength ratios from Jackson ratios
- Shoulder horizontal adduction/abduction and internal/external rotation is a scaled version of the Kentel. Strength was scaled using the Jackson-to-Kentel shoulder flex/ext ratios.
- Elbow extension/flexion and forearm pronation/supination. Elbow strength scaled from Kentel using the ratio of maximum isometric shoulder ext/flextion between Kentel and Jackson. Forearm pronation/supination scaled using the maximum torque strength ratio of wrist extension/flextion between Kentel and Jackson
- Wrist ext/flexion directly from Jackson, while the curves for ulnar and radial deviation have been scaled (using the maximum isometric torque ratios of wrist extension and flexion from both models) from Kentel et al.
- Lumbar-extension active-torque-angle-curve in extension comes from Raschke et al.
- Lumbar-extension passive-torque-angle-curve comes from Dolan et al.
- Lumbar-flexion
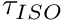 comes from Beimborn et al.'s observation that the strength ratio of back extensors to flextors at a flextion angle of zero is most often repored as 1.3:1.
comes from Beimborn et al.'s observation that the strength ratio of back extensors to flextors at a flextion angle of zero is most often repored as 1.3:1. - Lumbar-extension and flexion torque-velocity-curves have been estumated using the architecture of these two muscles. This update has been made because the force-velocity curve proposed by Raschke and Chaffin in Fig. 4 has such a low maximum angular velocity (60 dec/sec) that none of our optimal control simulations predicted lumbar flexion during movement. Using muscle archectural information for the erector spinae (ES) and rectus abdominus (RA), assuming both are made of slow twitch fibers the maximum angular velocites are 433 deg/sec and 1102 deg/sec about the constant assumed moment arms for the lumbar extensors and flexors respectively. The following architectural information was used:
- 7.1 cm :ES moment arm from Németh & Ohlsén
- 8.08 cm :ES optimal fiber length taken from a weighted PSCA average of the ES muscles from Christophy et al. Table 1
- 7.02
 :ES maximum angular velocity for slow twitch muscle from Ranatunga
:ES maximum angular velocity for slow twitch muscle from Ranatunga - 0.151 :ES tv at half max. angular velocity
- 10.9 cm :RA moment arm from Németh & Ohlsén
- 29.9 cm :RA optimal fiber length from Christophy et al. Table 1
- 7.02
 :RA maximum angular velocity for slow twitch muscle from Ranatunga
:RA maximum angular velocity for slow twitch muscle from Ranatunga - 0.151 :RA tv at half max. angular velocity for slow twitch muscle from Ranatunga
- Any passive curve that is not accompanied by a curve from the literature (see the plots for details) is an educated guess.
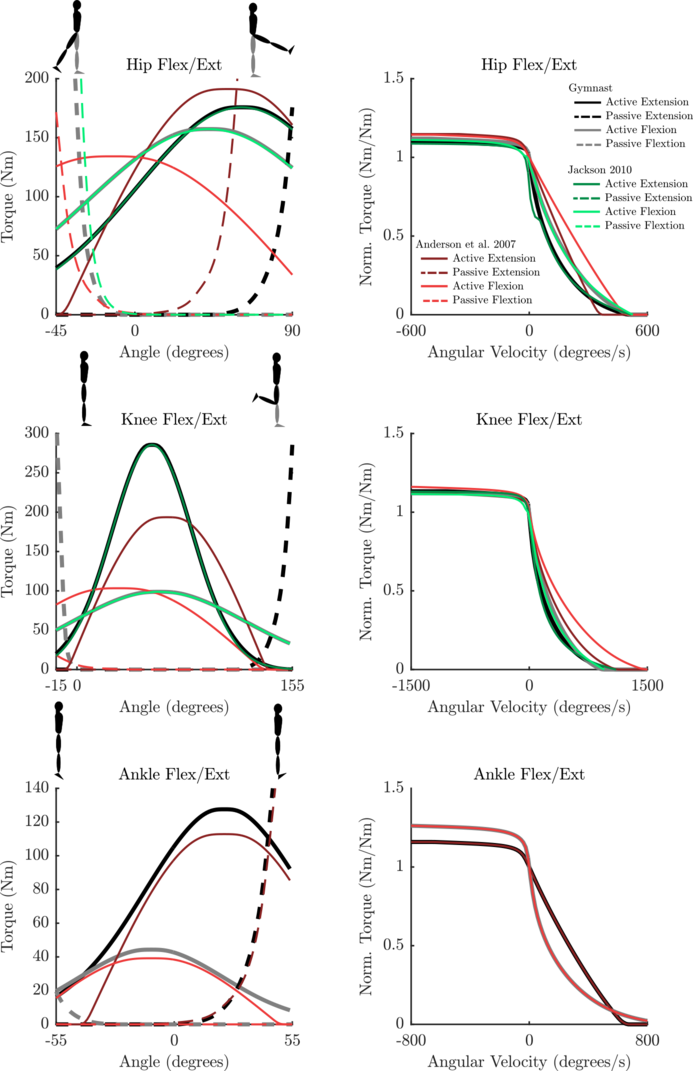
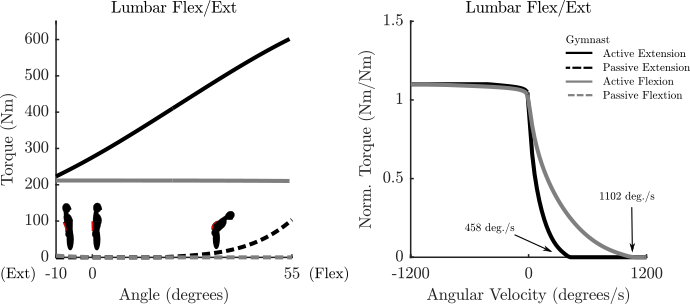
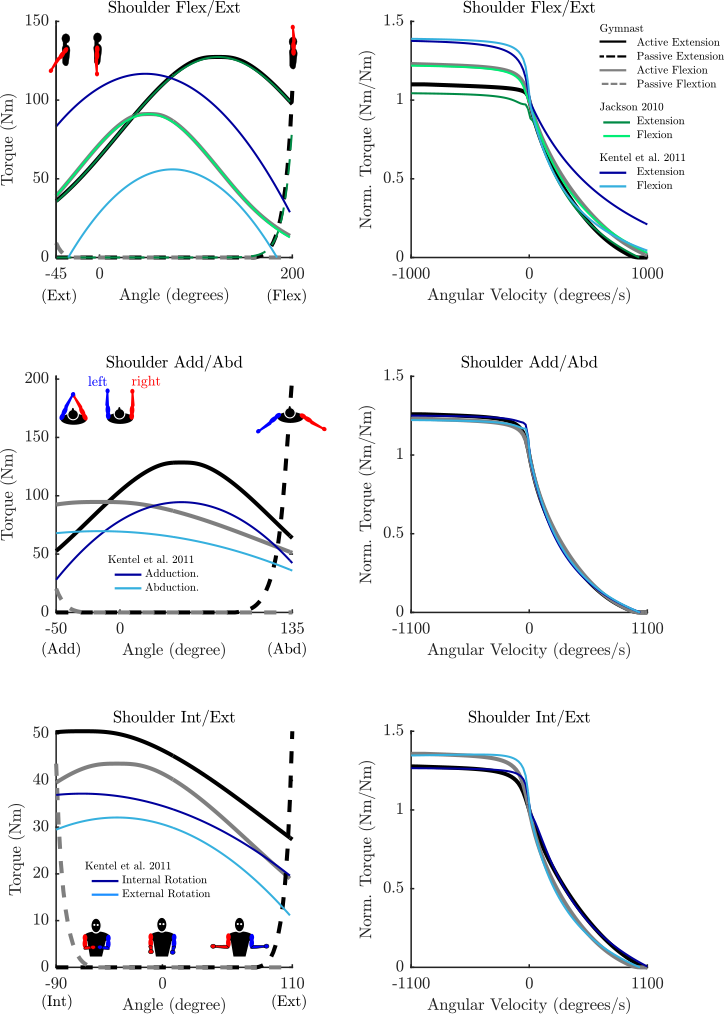
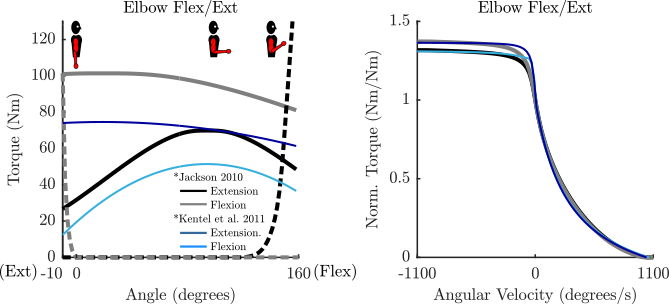
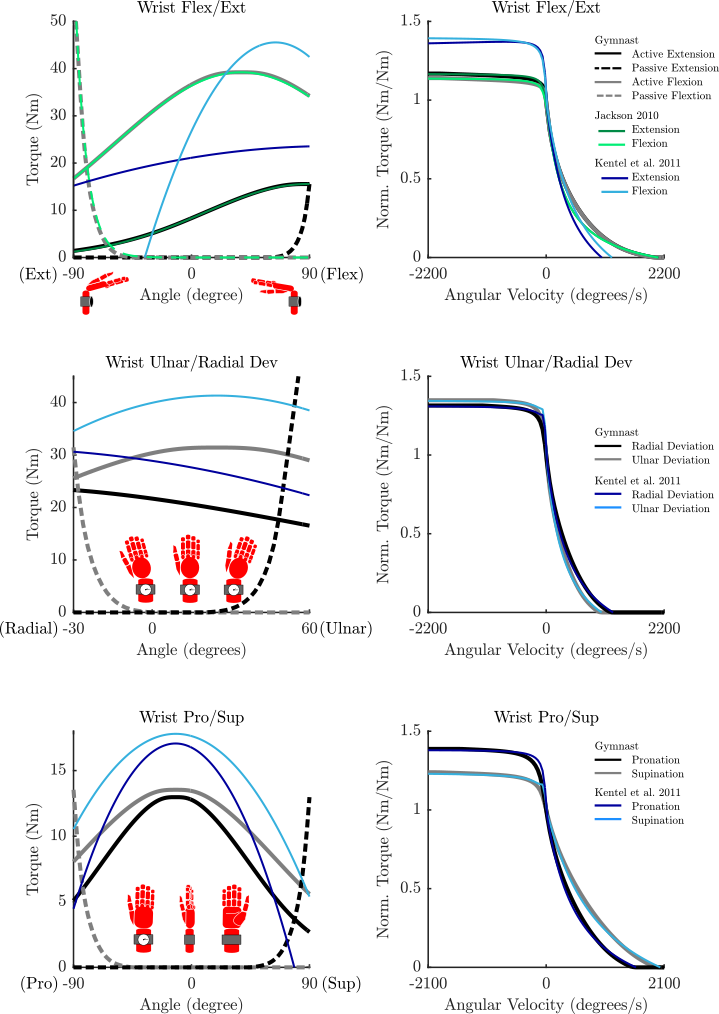
Parameterized Curves used here vs. Literature
The curves used in this implementation are 2nd order 2-dimensional Bezier curves. The curves described in Anderson et al., Jackson, Kentel were not directly used because they are not continuous to the second derivative (a requirement for most gradient based optimization routines). There are some other detailed differences that might be of interest:
- Anderson et al.'s torque-velocity curve tends to large negative values for fast eccentric contractions. This is in contrast to the literature which says that at large eccentric contractions the torque-velocity curve (or the force-velocity-curve) tends to a value between 1.0 and 1.4.
- Anderson et al.'s torque-velcity curve for ankle extension did not cross the x-axis on the concentric side of the curve. This would endow the plantar flexors with super-human abilities. This error has been corrected by fitting a Bezier curve to a Hill-type curve that passes through the point where
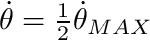
- Anderson et al.'s, Jackson, and Kentel et al. had discontinuities in the first derivative of the force velocity curve at
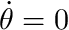 . While this follows Huxley's famous observations that the slope does discontinuously change at at
. While this follows Huxley's famous observations that the slope does discontinuously change at at 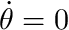 , this is not a phenomena that is not compatible with most optimal control formulations and thus this discontinuity is not present in the force velocity curves used in this model.
, this is not a phenomena that is not compatible with most optimal control formulations and thus this discontinuity is not present in the force velocity curves used in this model. - Anderson et al. and Kentel et al.'s active-torque-angle curves can achieve negative values - this is obviously undesirable as it will allow a muscle to push.
Kentel et al.'s activation inhibition function does not always cross 1.0 for
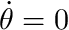 , which means that
, which means that 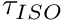 is not reached. This makes for a confusing model to use.
is not reached. This makes for a confusing model to use.Coordinate Mapping
Every author chose a particular convention for measuring the angles of the hip, knee, ankle joint, shoulder, elbow, wrist and lumbar — see the figure for details. These conventions have all been mapped to the one used in the illustrations. You will need to use the figure, your model, and the constructors appropriately so that
- the joint angle of your model is correctly mapped to the fiber angle of the Millard2016TorqueMuscle;
- the sign of the muscle's output torque matches the sign associated with your model.
To map from your model's joint coordinates to the joint coordines used in this model (see the figure in the description) the followinq equation is used at the torque level
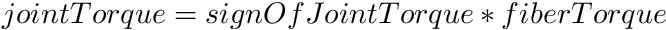
where fiberTorque is the torque produced by Anderson et al.'s curves, which is always positive. At the position level, the angles from your models joint angle to Anderson et al.'s joint angle (called fiberAngle) are mapped using
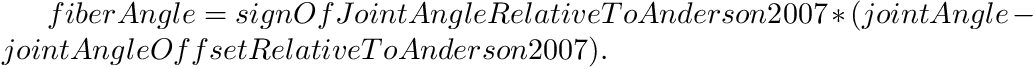
Internally the sign of the fiber velocity follows signOfJointTorque so that the signs of joint power and muscle power are consistent.
Strength Scaling: Anderson2007 Data Set
The leg strength (here we mean 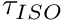 ) predicted by Anderson et al.'s curves should be taken as a good first approximation. While Anderson et al.'s data set is the most comprehensive in the literature, they only measured torques from active people: they did not include people at the extremes (both very weak, and very strong), nor did they include children. Finally, the torques produced by each subject were normalized by mSubjectMassInKg*subjectHeightInM*accelerationDueToGravity. Strength is a strange phenomena which is not nicely normalized by just these quantites, and so the strength predicted by Anderson et al.'s curves might not fit your subject even if they are represented in Anderson et al.'s data set.
) predicted by Anderson et al.'s curves should be taken as a good first approximation. While Anderson et al.'s data set is the most comprehensive in the literature, they only measured torques from active people: they did not include people at the extremes (both very weak, and very strong), nor did they include children. Finally, the torques produced by each subject were normalized by mSubjectMassInKg*subjectHeightInM*accelerationDueToGravity. Strength is a strange phenomena which is not nicely normalized by just these quantites, and so the strength predicted by Anderson et al.'s curves might not fit your subject even if they are represented in Anderson et al.'s data set.
Strength Scaling: Gymnast Data Set
The strength used in the Gymnast data set is fitted to an elite male gymnast. It goes without saying that an elite gymnast has strength proportions, and an absolute strength that are not typical. In the future it would be nice to have a function that could provide an educated guess about how to map Gymnast's strengths to that of another subject. For the moment I have no idea how to do this, nor am I aware of any works in the literature that can provide insight of how to do this. For now the whole-body Gymnast model should be viewed as being a representation of what is possible for a human, but not a typical human. At the present time the default strength settings of the Gymnast are not scaled by subject height, nor weight.
If you happen to know the maximum-isometric-active-torque (note this does not include the passive component) that your subject can produce, you can update the strength of the torque-muscle using the functions getMaximumActiveIsometricTorque(), and setMaximumActiveIsometricTorque().
Fitting to a Specific Subject
If you have recorded the motions and external forces acting on a subject during a movement of interest, you can make use of the TorqueMuscleFittingToolkit to adjust the MTG so that it is strong enough and flexible enough to allow a model of the subject to reproduce the experimental data. Please see the TorqueMuscleFittingToolkit class for details.
Limitations
This rigid-tendon torque muscle has some limitations that you should be aware of:
- There are no elastic tendons. That means that the mapping between the mechanical work done by this torque actuator will greatly differ from the positive mechanical work done by a torque actuator that includes an elastic tendon. This difference is greatest for those muscles with long tendons - namely the Achilles tendon in humans. If you are interested in fiber kinematics, fiber work, or metabolic energy consumption you cannot use this model especially for muscles that have long tendons.
- This model formulation predicts torque well, but does a poor job of predicting joint stiffness. In this model stiffness is given by the partial derivative of torque w.r.t. joint angle. Since the active-torque-angle curve fits a bell-shaped curve, it is possible to construct a torque muscle that has a region of physically impossible negative stiffness. Real muscle, in constrast, always has a positive stiffness even on the descending limb of the active-torque-angle curve (see Rassier et al. for details).
- Muscles that cross 2 joints (e.g. the hamstrings) produce coupled torques at both of those joints. In this model there is no coupling between joints. Furthermore, because of the lack of coupling the curves used here are only valid for the posture that Anderson et al., Jackson, and Kentel et al. used when they made their data collection. If you are interested in simulating postures that are very different from those described in by these authors then the results produced by this model should be treated as very rough.
- Because this is a joint-torque muscle, none of the joint contact forces predicted will come close to matching what is produced by line-type muscles. If you are interested in joint-contact forces you cannot use this model.
This simple model is a fast approximate means to constrain the joint torque developed in the body to something that is physiologically possible. That is it.
Units Although the figure in this description has angles in units of degrees, this is only to help intuitition: when using the model, use radians. This model uses MKS:
-Distance: m -Angles: radians -Angular velocity: radians/s -Mass: kg -Torque: Nm -Time: second -Power: Nm/second
Developer Notes
All of the numerical evaluations of this model take place in a few private stateless functions: -updateTorqueMuscleSummary -updTorqueMuscleSummaryCurveValues -updTorqueMuscleInfo -updInvertTorqueMuscleSummary If you want to change something fundamental about the mathematics of the model, then you have to update these functions.
References
- Anderson, D. E., Madigan, M. L., & Nussbaum, M. A. (2007). Maximum voluntary joint torque as a function of joint angle and angular velocity: model development and application to the lower limb. Journal of biomechanics, 40(14), 3105-3113.
- Beimborn, D. S., & Morrissey, M. C. (1988). A review of the literature related to trunk muscle performance. Spine, 13(6), 655-660.
- Christophy, M., Senan, N. A. F., Lotz, J. C., & O’Reilly, O. M. (2012). A musculoskeletal model for the lumbar spine. Biomechanics and Modeling in Mechanobiology, 11(1-2), 19-34.
- Dolan, P., A. F. Mannion, and M. A. Adams. Passive tissues help the back muscles to generate extensor moments during lifting. Journal of Biomechanics 27, no. 8 (1994): 1077-1085.
- Jackson, M.I. (2010). The mechanics of the Table Contact Phase of Gymnastics Vaulting. Doctoral Thesis, Loughborough University.
- Kentel, B.B., King, M.A., & Mitchell, S.R. (2011). Evaluation of a subject-specific torque-driven computer simulation model of one-handed tennis backhand ground strokes. Journal of Applied Biomechanics, 27(4),345-354.
- Millard, M., Uchida, T., Seth, A., & Delp, S. L. (2013). Flexing computational muscle: modeling and simulation of musculotendon dynamics. Journal of biomechanical engineering, 135(2), 021005.
- Németh, G., & Ohlsén, H. (1986). Moment arm lengths of trunk muscles to the lumbosacral joint obtained in vivo with computed tomography. Spine, 11(2), 158-160.
- Ranatunga, K. W. (1984). The force-velocity relation of rat fast-and slow-twitch muscles examined at different temperatures. The Journal of Physiology, 351, 517.
- Raschke, U., & Chaffin, D. B. (1996). Support for a linear length-tension relation of the torso extensor muscles: an investigation of the length and velocity EMG-force relationships. Journal of biomechanics, 29(12), 1597-1604.
- Rassier, D. E., Herzog, W., Wakeling, J., & Syme, D. A. (2003). Stretch-induced, steady-state force enhancement in single skeletal muscle fibers exceeds the isometric force at optimum fiber length. Journal of biomechanics, 36(9), 1309-1316.
Definition at line 922 of file Millard2016TorqueMuscle.h.
Constructor & Destructor Documentation
◆ Millard2016TorqueMuscle() [1/2]
Default constructor, which for the moment does nothing. Calling any of the models functions after the default construction will result in a runtime error.
◆ Millard2016TorqueMuscle() [2/2]
| Millard2016TorqueMuscle | ( | DataSet::item | dataSet, |
| const SubjectInformation & | subjectInfo, | ||
| int | jointTorque, | ||
| double | jointAngleOffsetRelativeToDoxygenFigures, | ||
| double | signOfJointAngleRelativeToDoxygenFigures, | ||
| double | signOfJointTorqueToDoxygenFigures, | ||
| const std::string & | name | ||
| ) |
This constructor allows you to easily access the large table of built-in torque muscle coefficients to create a torque muscle that best represents the joint of interest.
Note: directions This constructs a single joint-torque muscle: it can only generate torque in one direction. If you want to generate a torque in two directions, you need 2 torque muscles.
Note: signs and offsets
All of the angles in these models are defined anatomically. You will need to set a series of variables to correctly map from your model's joint coordinates and sign conventions to that of the models: jointAngleOffsetRelativeToDoxygenFigures, signOfJointAngleRelativeToDoxygenFigures, signOfJointTorqueToDoxygenFigures. Also note that due to the anatomical angle definitions some left and right handed joints will require different signs. This will be true for internal/external rotation at the shoulder, horizontal adduction/abduction at the shoulder, ulnar/radial deviation at the wrist, pronation/supination of the wrist, and others as the list of directions grows.
Note: blending variables
When this constructor is called the blending variables for the active-torque-angle-curve, torque-angular-velocity-curve, and passive-torque-angle-curve are set to 0.
- Parameters
-
dataSet The desired source of joint torque coefficients. Use the DataSet structure to choose the desired data set (e.g. DataSet::Anderson2007, or DataSet::Gymnast) subjectInfo A struct that contains metadata about the subject which is used to scale the maximum torque of the torque muscle. jointTorque Select the joint and torque direction of interest. Use the struct for each data set to choose a joint-torque-direction that is in the set (e.g. Anderson2007::HipExtension, or Gymnast::ShoulderHorizontalAdduction) jointAngleOffsetRelativeToDoxygenFigures Offset angle between your model's joints and the reference figures in class description. signOfJointAngleRelativeToDoxygenFigures The sign convention that converts your model's joint angles to the angles used in the reference figures. signOfJointTorqueToDoxygenFigures The sign that maps fiberTorque from Anderson's model (which is always positive) to the correctly signed joint torque for your model. name The name of the muscle. This is needed to do useful things like provide error messages that are human readable.
- Exceptions
-
abort() when - The combination of dataSet, gender, joint, and jointDirection does not correspond to a valid entry
- mSubjectHeightInMeters <= 0
- mSubjectMassInKg <= 0
- abs(signOfJointTorque)-1 > epsilon
Member Function Documentation
◆ calcActivation()
| void calcActivation | ( | double | jointAngle, |
| double | jointAngularVelocity, | ||
| double | jointTorque, | ||
| TorqueMuscleSummary & | updTorqueMuscleSummaryStruct | ||
| ) | const |
This function will calculate the muscle activation needed to generate the joint torque. This function is useful in the process of fitting the strength of a model to inverse-dynamics data. The value for activation is arrived at by evaluating this equation
![\[ \mathbf{a} = \dfrac{ \dfrac{\tau (\mathbf{a}, \theta,\dot{\theta})}{ \tau_{ISO}} - \mathbf{t}_P(1- \beta (\dot{\theta}/\dot{\theta}_{MAX}))}{ \mathbf{t}_A(\theta) \mathbf{t}_V(\dot{\theta}/\dot{\theta}_{MAX})}. \]](../../form_191.png)
If the passive forces at the desired angle actually exceed the desired torque, a negative value for activation will be returned. If you see this it just means the passive-torque-angle curve of this muscle needs to be adjusted.
Note An activation value of 0 is passed out if the sign of the desired jointTorque differs from the sign of the joint torque that this muscle can produce.
- Parameters
-
jointAngle (radians) jointAngularVelocity (radians/sec) jointTorque (Nm). updTorqueMuscleSummaryStruct TorqueMuscleSummary struct which contains the calculated activation along with all of the internal parameters of the muscle so that you can understand why the activation value takes the value that it does. See the TorqueMuscleSummary (in this file) for details.
◆ calcBlendedCurveDerivative()
|
private |
This function will calculate the value of a blended smooth segmented curve 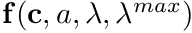 , where
, where  is a smooth-segmented-curve
is a smooth-segmented-curve
![\[ \mathbf{f}(a,\lambda,\lambda^{max},\mathbf{c}()) = \mathbf{c}(a) (1-\lambda) + \lambda\lambda^{max} \]](../../form_205.png)
Note At the moment this function only returns first derivatives.
- Parameters
-
curveArgument [ 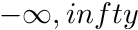 ] the argument
] the argument 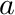 to the smooth-segmented-curve
to the smooth-segmented-curve 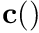 . Units: depend on the curve.
. Units: depend on the curve.blendingVariable [0-1]. the value of 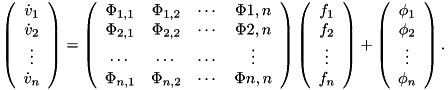 the blending variable. Unitless
the blending variable. UnitlessmaximumBlendingValue the scalar value the blended function tends to with a blending variable 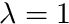 .
.derivativeOrderArgument [0,1]. The order of the derivative of the SmoothSegmentedFunction w.r.t. its scalar input argument. derivativeOrderBlendingVariable [0,1]. This parameter is set to choose the derivative order with respect to the blending variable. curve the SmoothSegmentedCurve to evaluate.
- Returns
- the value of the blended curve or its derivative
◆ calcFiberAngle()
|
private |
◆ calcFiberAngleGivenNormalizedPassiveTorque()
|
private |
This function will calculate the angle at which the blendable passive torque angle curve develops a desired torque. Note that if the blendingVariable is set to 1.0 then the curve is y = 0, and a NaN will be returned.
- Parameters
-
normPassiveFiberTorque the normalized passive fiber torque blendingVariable A variable between [0,1] where 0 is equivalent to using the default passive-torque-angle curve and 1 will set this curve to 0. Any value between 0-1 will smoothly interpolate between these extremes. passiveTorqueAngleCurveOffset the angular shift of the passive-torque-angle curve.
- Returns
- fiber angle in radians at which the blendable passive curve developes the desired curve.
◆ calcFiberAngularVelocity()
|
private |
◆ calcInverseBlendedCurveValue()
|
private |
This function will calculate the inverse value of a blended smooth segmented curve, that is 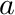 , in the function
, in the function
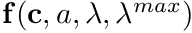 , where
, where  is a smooth-segmented-curve
is a smooth-segmented-curve
![\[ \mathbf{f}(a,\lambda,\lambda^{max},\mathbf{c}()) = \mathbf{c}(a) (1-\lambda) + \lambda\lambda^{max} \]](../../form_205.png)
Note At the moment this function only returns first derivatives.
- Parameters
-
blendedCurveValue the value of 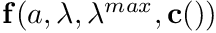
argGuess an initial guess for the value of 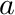 , which is necessary as not all blended functions are monotonic.
, which is necessary as not all blended functions are monotonic.blendingVariable [0-1]. the value of 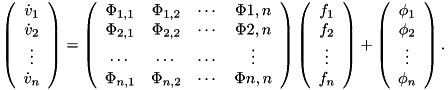 the blending variable. Unitless
the blending variable. UnitlessmaximumBlendingValue the scalar value the blended function tends to with a blending variable 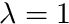 .
.curve the SmoothSegmentedCurve to evaluate.
- Returns
- the value of the argument
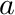 which will evaluate to the given value of
which will evaluate to the given value of 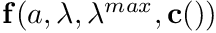
◆ calcJointAngle()
|
private |
◆ calcJointAngularVelocity()
|
private |
◆ calcJointTorque()
| double calcJointTorque | ( | double | jointAngle, |
| double | jointAngularVelocity, | ||
| double | activation | ||
| ) | const |
Calculates the signed joint torque developed by the muscle. Note that the signs that are needed to map from the native curves to those of your specific model are set when the muscle is constructed.
![\[ \tau_{M} = \pm \tau_{ISO}( \mathbf{a} \, \mathbf{t}_A(\theta) \mathbf{t}_V(\dot{\theta}/\dot{\theta}_{MAX}) +\mathbf{t}_P(1- \beta (\dot{\theta}/\dot{\theta}_{MAX})) \, ) \]](../../form_190.png)
- Parameters
-
jointAngle (radians) jointAngularVelocity (radians/sec) activation the percentage of the muscle that is turned on [0-1]. This function allows activations to be outside [0,1], because this is useful during the intermediate solutions of an optimization run. However, you must ensure after the fact that your activations fall within a bound of [0,1].
- Returns
- torque developed by the muscle in (Nm).
◆ calcMaximumActiveIsometricTorqueScalingFactor()
| double calcMaximumActiveIsometricTorqueScalingFactor | ( | double | jointAngle, |
| double | jointAngularVelocity, | ||
| double | activation, | ||
| double | jointTorque | ||
| ) | const |
This function will compute the scaling factor 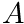 that would be needed in order for this muscle to generate a specific joint torque
that would be needed in order for this muscle to generate a specific joint torque 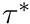 at a specific activation level.
at a specific activation level.
![\[ A = \dfrac{\tau^*}{ \tau_{ISO}( \mathbf{a} \, \mathbf{t}_A(\theta) \mathbf{t}_V(\dot{\theta}/\dot{\theta}_{MAX}) +\mathbf{t}_P(1- \beta (\dot{\theta}/\dot{\theta}_{MAX})) \, )} \]](../../form_194.png)
This function is useful in the process of determining how to much to scale the strength of a default model to match inverse-dynamics data of a subject. In contrast to calcActivation, if the passive forces of the muscle exceed the desired joint torque the resulting scale factor will drop.
- Parameters
-
jointAngle (radians) jointAngularVelocity (radians/sec) jointTorque (Nm) activation
- Returns
- scaleFactor: The scale that would make the current muscle produce exactly the desired torque at the specfied angle and angular velocity. Note: a value of 0 is passed out if the sign of the desired jointTorque differs from the sign of the joint torque that this muscle can produce.
◆ calcTorqueMuscleDataFeatures()
|
private |
◆ calcTorqueMuscleInfo()
| void calcTorqueMuscleInfo | ( | double | jointAngle, |
| double | jointAngularVelocity, | ||
| double | activation, | ||
| TorqueMuscleInfo & | updTorqueMuscleInfoStruct | ||
| ) | const |
Calculates a large number of internal quantities of the torque muscle ranging from the values of the muscle's components, the stiffness of the muscle, and its power output. See the struct TorqueMuscleInfo (in this file) for details.
- Parameters
-
jointAngle (radians) jointAngularVelocity (radians/sec) activation the percentage of the muscle that is turned on [0-1]. This function allows activations to be outside [0,1], because this is useful during the intermediate solutions of an optimization run. However, you must ensure after the fact that your activations fall within a bound of [0,1]. updTorqueMuscleInfoStruct A torque muscle struct
◆ fitPassiveCurveAngleOffset()
| void fitPassiveCurveAngleOffset | ( | double | jointAngle, |
| double | passiveFiberTorque | ||
| ) |
This function solves for the passive curve angle offset so that at the specified jointAngle the passive curve develops the specified passiveTorque
- Parameters
-
jointAngle the target joint angle in radians passiveFiberTorque the target passive joint torque in Nm.
- Exceptions
-
abort if passiveTorque < sqrt(eps)
◆ fitPassiveTorqueScale()
| void fitPassiveTorqueScale | ( | double | jointAngle, |
| double | passiveFiberTorque | ||
| ) |
This function iteratively solves for the scaling so that at the specified jointAngle the passive curve develops the specified passiveTorque
- Parameters
-
jointAngle the target joint angle in radians passiveFiberTorque the target passive joint torque in Nm.
- Exceptions
-
abort if jointAngle is not in the domain of the curve abort if passiveFiberTorque < sqrt(eps)
◆ getActiveTorqueAngleCurve()
| const RigidBodyDynamics::Addons::Geometry::SmoothSegmentedFunction & getActiveTorqueAngleCurve | ( | ) | const |
- Returns
- the SmoothSegmentedFunction represents the active-torque-angle curve profile.
◆ getActiveTorqueAngleCurveAngleScaling()
| double getActiveTorqueAngleCurveAngleScaling | ( | ) | const |
- Returns
- the scaling parameter that is applied to the domain of the active-torque-angle curve. Note that a value greater than 1 means that the curve has been stretched.
◆ getActiveTorqueAngleCurveBlendingVariable()
| double getActiveTorqueAngleCurveBlendingVariable | ( | ) | const |
- Returns
- Value of
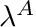 that is currently in use.
that is currently in use.
◆ getActiveTorqueAngleCurveWidth()
| double getActiveTorqueAngleCurveWidth | ( | ) | const |
- Returns
- the width of the active torque angle curve. Note that this will return the angular width of the non-blended curve. Angle is in radians.
◆ getAgeGroup()
| AgeGroupSet::item getAgeGroup | ( | ) |
◆ getDataSet()
| DataSet::item getDataSet | ( | ) |
◆ getGender()
| GenderSet::item getGender | ( | ) |
◆ getJointAngleAtMaximumActiveIsometricTorque()
| double getJointAngleAtMaximumActiveIsometricTorque | ( | ) | const |
- Returns
- the joint angle at which the normalized active-torque-angle curve peaks at its maximum value of 1.0. Angle is in radians
◆ getJointAngleAtOneNormalizedPassiveIsometricTorque()
| double getJointAngleAtOneNormalizedPassiveIsometricTorque | ( | ) | const |
- Returns
- the joint angle at which the normalized passive-torque-angle curve reaches a value of 1.0. If this curve never reaches a value of 1.0 (because it is flat, or the mPassiveTorqueScale has been set to 0) a value of std::numeric_limits<double>::signaling_NaN() is returned. Use the std function isfinite to test if a signaling_NaN has been returned. Angle is in radians
◆ getJointAngleAtSmallestNormalizedPassiveIsometricTorque()
| double getJointAngleAtSmallestNormalizedPassiveIsometricTorque | ( | ) | const |
- Returns
- the joint angle at which the normalized passive-torque-angle curve reaches its minimum value Angle is in radians
◆ getJointAngleOffset()
| double getJointAngleOffset | ( | ) | const |
- Returns
- the offset angle between the model's joint and the figures in the class description (rad)
◆ getJointAngleSign()
| double getJointAngleSign | ( | ) | const |
- Returns
- the sign of the angle sign relative to the figures in the class description (+/- 1)
◆ getJointTorque()
| JointTorqueSet::item getJointTorque | ( | ) |
◆ getJointTorqueSign()
| double getJointTorqueSign | ( | ) | const |
- Returns
- the sign of the joint torque (+/- 1)
◆ getMaximumActiveIsometricTorque()
| double getMaximumActiveIsometricTorque | ( | ) | const |
- Returns
- the maximum-active-isometric torque that this muscle can produce in Nm.
◆ getMaximumConcentricJointAngularVelocity()
| double getMaximumConcentricJointAngularVelocity | ( | ) | const |
- Returns
- the maximum concentric angular velocity in radians/sec. This scalar positive quantity corresponds to the angular speed at which the torque-velocity curve crosses zero.
◆ getName()
| std::string getName | ( | ) |
◆ getNormalizedDampingCoefficient()
| double getNormalizedDampingCoefficient | ( | ) | const |
- Returns
- The maximum normalized damping term
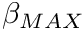 term in the torque muscle model. See class description for details.
term in the torque muscle model. See class description for details.
◆ getPassiveCurveAngleOffset()
| double getPassiveCurveAngleOffset | ( | ) | const |
- Returns
- the angle
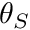 that the passive curve has been shifted. Note that the sign convention is in the sense of the fiber angle, not in the sense of the joint angle. (radians).
that the passive curve has been shifted. Note that the sign convention is in the sense of the fiber angle, not in the sense of the joint angle. (radians).
◆ getPassiveTorqueAngleCurve()
| const RigidBodyDynamics::Addons::Geometry::SmoothSegmentedFunction & getPassiveTorqueAngleCurve | ( | ) | const |
- Returns
- the SmoothSegmentedFunction that represents the passive-torque-angle curve.
◆ getPassiveTorqueAngleCurveBlendingVariable()
| double getPassiveTorqueAngleCurveBlendingVariable | ( | ) | const |
- Returns
- Value of
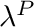 that is currently in use.
that is currently in use.
◆ getPassiveTorqueScale()
| double getPassiveTorqueScale | ( | ) | const |
- Returns
- the passive-torque-scale
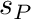 that is applied to the passive-torque-curve.
that is applied to the passive-torque-curve.
◆ getSubjectHeight()
| double getSubjectHeight | ( | ) |
◆ getSubjectMass()
| double getSubjectMass | ( | ) |
◆ getTorqueAngularVelocityCurve()
| const RigidBodyDynamics::Addons::Geometry::SmoothSegmentedFunction & getTorqueAngularVelocityCurve | ( | ) | const |
- Returns
- the SmoothSegmentedFunction the represents the torque-angular-velocity curve.
◆ getTorqueAngularVelocityCurveBlendingVariable()
| double getTorqueAngularVelocityCurveBlendingVariable | ( | ) | const |
- Returns
- Value of
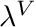 that is currently in use.
that is currently in use.
◆ getTorqueVelocityMultiplierAtHalfOmegaMax()
| double getTorqueVelocityMultiplierAtHalfOmegaMax | ( | ) | const |
◆ printJointTorqueProfileToFile()
| void printJointTorqueProfileToFile | ( | const std::string & | path, |
| const std::string & | fileNameWithoutExtension, | ||
| int | numberOfSamplePoints | ||
| ) |
Prints 2 csv files:
'fileName' + '_variableLengthfixedVelocity': All of the fields in TorqueMuscleInfo are recorded to file as the jointAngle varies but the jointAngularVelocity is zero.
-#'fileName' + '_fixedLengthVariableVelocity': All of the fields in TorqueMuscleInfo are recorded to file as the jointAngle is fixed but the jointAngularVelocity varies.Each column has a header, so that you can tell what each piece of data means.
- Parameters
-
path the path to the destination folder. Don't put an '\' on the end. fileNameWithoutExtension the name of the file, but without an extension.
numberOfSamplePoints the number of sample points to use in the files.
◆ setActiveTorqueAngleCurveAngleScaling()
| void setActiveTorqueAngleCurveAngleScaling | ( | double | angleScaling | ) |
- Parameters
-
angleScaling Scale the domain of the active torque angle curve. This scaling is applied so that the angle at which the curve peaks remains unchanged. Note that an angleScaling > 1 means that the typically bell-shaped curve will be wider.
◆ setActiveTorqueAngleCurveBlendingVariable()
| void setActiveTorqueAngleCurveBlendingVariable | ( | double | blendingVariable | ) |
- Parameters
-
blendingVariable [0-1] The desired value of 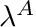 . Note at
. Note at 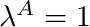 the curve is a horizontal line with a value of 1.
the curve is a horizontal line with a value of 1.
◆ setFittedParameters()
| void setFittedParameters | ( | const TorqueMuscleParameterFittingData & | fittedParameters | ) |
- Parameters
-
fittedParameters the structure returned after one of the optimization-based fitting functions from TorqueMuscleFittingToolkit has been called. If the fitting was successful, calling this function will update all of the adjusted parameters.
◆ setMaximumActiveIsometricTorque()
| void setMaximumActiveIsometricTorque | ( | double | maxIsometricTorque | ) |
Sets the strength of the muscle to match a desired value.
- Parameters
-
maxIsometricTorque The desired maximum-active-isometric torque of the muscle (Nm)
◆ setMaximumConcentricJointAngularVelocity()
| void setMaximumConcentricJointAngularVelocity | ( | double | maxAngularVelocity | ) |
- Parameters
-
maxAngularVelocity the maximum concentric joint angular velocity magnitude in radians/sec. This scalar quantity is not signed and must be greater than 0.
◆ setName()
| void setName | ( | std::string & | name | ) |
◆ setNormalizedDampingCoefficient()
| void setNormalizedDampingCoefficient | ( | double | beta | ) |
- Parameters
-
beta -t normalized damping term 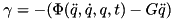 term in the torque muscle model. See class description for details.
term in the torque muscle model. See class description for details.
- Exceptions
-
abort() if beta < 0
◆ setPassiveCurveAngleOffset()
| void setPassiveCurveAngleOffset | ( | double | passiveCurveAngleOffsetVal | ) |
- Parameters
-
passiveCurveAngleOffsetVal the angle 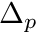 that the passive curve should be shifted. Note that the sign convention is in the sense of the fiber angle, not in the sense of the joint angle. Angles in radians
that the passive curve should be shifted. Note that the sign convention is in the sense of the fiber angle, not in the sense of the joint angle. Angles in radians
◆ setPassiveTorqueAngleCurveBlendingVariable()
| void setPassiveTorqueAngleCurveBlendingVariable | ( | double | blendingVariable | ) |
- Parameters
-
blendingVariable [0-1] The desired value of 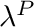 . Note at
. Note at 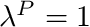 the curve is a horizontal line with a value of 0.
the curve is a horizontal line with a value of 0.
◆ setPassiveTorqueScale()
| void setPassiveTorqueScale | ( | double | passiveTorqueScale | ) |
Sets the scaling of the passive-joint-torques. By default this scale is one.
- Parameters
-
passiveTorqueScale The scale 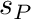 applied to the passive-joint-torque curve (unitless)
applied to the passive-joint-torque curve (unitless)
◆ setTorqueAngularVelocityCurveBlendingVariable()
| void setTorqueAngularVelocityCurveBlendingVariable | ( | double | blendingVariable | ) |
- Parameters
-
blendingVariable [0-1] The desired value of 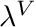 . Note at
. Note at 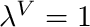 the curve is a horizontal line with a value of 1.
the curve is a horizontal line with a value of 1.
◆ setTorqueVelocityMultiplierAtHalfOmegaMax()
| void setTorqueVelocityMultiplierAtHalfOmegaMax | ( | double | tvAtHalfOmegaMax | ) |
◆ updateTorqueMuscleCurves()
|
private |
◆ updInvertTorqueMuscleSummary()
|
private |
This function evaluates all of the quantities needed to compute the activation of the muscle given its torque. In this case the activation assigned to the TorqueMuscleSummary structure is allowed to go outside of the bounds [0,1].
- Parameters
-
jointTorque the torque generated by the muscle at the joint (Nm) jointAngle the angle of the joint(radians) jointAngularVelocity the angular velocity of the joint ((rad/s)) activeTorqueAngleBlendingVariable the blending variable associated with the active-torque-angle curve passiveTorqueAngleBlendingVariable the blending variable associated with the passive-torque-angle curve activeTorqueAngularVelocityBlendingVariable the blending variable associated with the torque-angular velocity curve.
activeTorqueAngleAngleScaling the scaling of the angle. By default this is set to 1.0, but if desired, the active torque angle curve can be re-scaled to make it wider or narrower. This scaling is done such that the angular location of peak active-torque-angle multiplier does not change. activeTorqueAngleAtOneNormTorque the angle at which the torque-angle curve hits a normalized value of 1.0. passiveTorqueAngleCurveOffset the angular shift of the passive-torque-angle curve. maxAngularVelocity the maximum angular velocity of the muscle - used to normalize the angular velocity prior to evaluating the force-velocity curve. maxActiveIsometricTorque the maximum isometric torque updTms the torque-muscle-summary structure to be updated.
◆ updTorqueMuscleInfo()
|
private |
This function evaluates the curves values and derivatives in the TorqueMuscleInfo structue that are functions of fiberAngle and the normalized fiber angular velocity.
- Parameters
-
activation the activation of the muscle [0-1] jointAngle the angle of the joint(radians) jointAngularVelocity the angular velocity of the joint ((rad/s)) activeTorqueAngleBlendingVariable the blending variable associated with the active-torque-angle curve passiveTorqueAngleBlendingVariable the blending variable associated with the passive-torque-angle curve activeTorqueAngularVelocityBlendingVariable the blending variable associated with the torque-angular velocity curve. activeTorqueAngleAngleScaling the scaling of the angle. By default this is set to 1.0, but if desired, the active torque angle curve can be re-scaled to make it wider or narrower. This scaling is done such that the angular location of peak active-torque-angle multiplier does not change. activeTorqueAngleAtOneNormTorque the angle at which the torque-angle curve hits a normalized value of 1.0. passiveTorqueAngleCurveOffset the angular shift of the passive-torque-angle curve. maxAngularVelocity the maximum angular velocity of the muscle - used to normalize the angular velocity prior to evaluating the force-velocity curve. maxActiveIsometricTorque the maximum active isometric torque the muscle can generate updTmi the torque-muscle-info structure to be updated.
◆ updTorqueMuscleSummary()
|
private |
This function evaluates all of the quantities needed to compute the torque the muscle is generating but nothing extra.
- Parameters
-
activation the activation of the muscle which can range from 0-1. jointAngle the angle of the joint(radians) jointAngularVelocity the angular velocity of the joint ((rad/s)) activeTorqueAngleBlendingVariable the blending variable associated with the active-torque-angle curve passiveTorqueAngleBlendingVariable the blending variable associated with the passive-torque-angle curve activeTorqueAngularVelocityBlendingVariable the blending variable associated with the torque-angular velocity curve. activeTorqueAngleAngleScaling the scaling of the angle. By default this is set to 1.0, but if desired, the active torque angle curve can be re-scaled to make it wider or narrower. This scaling is done such that the angular location of peak active-torque-angle multiplier does not change. activeTorqueAngleAtOneNormTorque the angle at which the torque-angle curve hits a normalized value of 1.0. passiveTorqueAngleCurveOffset the angular shift of the passive-torque-angle curve. maxAngularVelocity the maximum angular velocity of the muscle - used to normalize the angular velocity prior to evaluating the force-velocity curve. maxActiveIsometricTorque the maximum isometric torque updTms the torque-muscle-summary structure to be updated.
◆ updTorqueMuscleSummaryCurveValues()
|
private |
This function evaluates the curves values that are a part of the torque-muscle-summary structure.
- Parameters
-
fiberAngle the angle of the muscle fiber (radians) normFiberAngularVelocity the normalized angular velocity of the fiber ((rad/s)/(rad/s)) activeTorqueAngleBlendingVariable the blending variable associated with the active-torque-angle curve passiveTorqueAngleBlendingVariable the blending variable associated with the passive-torque-angle curve activeTorqueAngularVelocityBlendingVariable the blending variable associated with the torque-angular velocity curve. activeTorqueAngleAngleScaling the scaling of the angle. By default this is set to 1.0, but if desired, the active torque angle curve can be re-scaled to make it wider or narrower. This scaling is done such that the angular location of peak active-torque-angle multiplier does not change. activeTorqueAngleAtOneNormTorque the angle at which the torque-angle curve hits a normalized value of 1.0. passiveTorqueAngleCurveOffset the angular shift of the passive-torque-angle curve. updTms the torque-muscle-summary structure to be updated.
Friends And Related Function Documentation
◆ FitTorqueMuscleParameters
|
friend |
Definition at line 925 of file Millard2016TorqueMuscle.h.
◆ TorqueMuscleFittingToolkit
|
friend |
Definition at line 924 of file Millard2016TorqueMuscle.h.
Field Documentation
◆ Anderson2007Table3Mean
|
staticprivate |
Definition at line 1527 of file Millard2016TorqueMuscle.h.
◆ Anderson2007Table3Std
|
staticprivate |
Definition at line 1528 of file Millard2016TorqueMuscle.h.
◆ GymnastWholeBody
|
staticprivate |
Definition at line 1529 of file Millard2016TorqueMuscle.h.
◆ mAgeGroup
|
private |
Definition at line 1487 of file Millard2016TorqueMuscle.h.
◆ mAnderson2007b1k1b2k2
|
private |
Definition at line 1482 of file Millard2016TorqueMuscle.h.
◆ mAnderson2007c1c2c3c4c5c6
|
private |
Definition at line 1481 of file Millard2016TorqueMuscle.h.
◆ mAngleAtOneNormActiveTorque
|
private |
Definition at line 1497 of file Millard2016TorqueMuscle.h.
◆ mAngleAtOneNormPassiveTorque
|
private |
Definition at line 1500 of file Millard2016TorqueMuscle.h.
◆ mAngleAtSmallestNormPassiveTorque
|
private |
Definition at line 1501 of file Millard2016TorqueMuscle.h.
◆ mAngleOffset
|
private |
Definition at line 1514 of file Millard2016TorqueMuscle.h.
◆ mBetaMax
|
private |
Definition at line 1505 of file Millard2016TorqueMuscle.h.
◆ mDataSet
|
private |
Definition at line 1485 of file Millard2016TorqueMuscle.h.
◆ mGender
|
private |
Definition at line 1486 of file Millard2016TorqueMuscle.h.
◆ mGymnastParams
|
private |
Definition at line 1483 of file Millard2016TorqueMuscle.h.
◆ mJointTorque
|
private |
Definition at line 1488 of file Millard2016TorqueMuscle.h.
◆ mMaxActiveIsometricTorque
|
private |
Definition at line 1496 of file Millard2016TorqueMuscle.h.
◆ mMuscleCurvesAreDirty
|
private |
Definition at line 1456 of file Millard2016TorqueMuscle.h.
◆ mMuscleName
|
private |
Definition at line 1516 of file Millard2016TorqueMuscle.h.
◆ mOmegaMax
|
private |
Definition at line 1499 of file Millard2016TorqueMuscle.h.
◆ mPassiveCurveAngleOffset
|
private |
Definition at line 1503 of file Millard2016TorqueMuscle.h.
◆ mPassiveTorqueScale
|
private |
Definition at line 1502 of file Millard2016TorqueMuscle.h.
◆ mScaleFactorAnderson2007
|
private |
Definition at line 1509 of file Millard2016TorqueMuscle.h.
◆ mSignOfConcentricAnglularVelocity
|
private |
Definition at line 1512 of file Millard2016TorqueMuscle.h.
◆ mSignOfJointAngle
|
private |
Definition at line 1511 of file Millard2016TorqueMuscle.h.
◆ mSignOfJointTorque
|
private |
Definition at line 1513 of file Millard2016TorqueMuscle.h.
◆ mSubjectHeightInMeters
|
private |
Definition at line 1507 of file Millard2016TorqueMuscle.h.
◆ mSubjectMassInKg
|
private |
Definition at line 1508 of file Millard2016TorqueMuscle.h.
◆ mTaAngleScaling
|
private |
Definition at line 1498 of file Millard2016TorqueMuscle.h.
◆ mTaCurve
|
private |
Definition at line 1462 of file Millard2016TorqueMuscle.h.
◆ mTaLambda
|
private |
Definition at line 1463 of file Millard2016TorqueMuscle.h.
◆ mTaLambdaMax
|
staticprivate |
Definition at line 1464 of file Millard2016TorqueMuscle.h.
◆ mTmInfo
|
private |
Definition at line 1458 of file Millard2016TorqueMuscle.h.
◆ mTmSummary
|
private |
Definition at line 1459 of file Millard2016TorqueMuscle.h.
◆ mTorqueVelocityMultiplierAtHalfOmegaMax
|
private |
Definition at line 1494 of file Millard2016TorqueMuscle.h.
◆ mTpCurve
|
private |
Definition at line 1466 of file Millard2016TorqueMuscle.h.
◆ mTpLambda
|
private |
Definition at line 1467 of file Millard2016TorqueMuscle.h.
◆ mTpLambdaMax
|
staticprivate |
Definition at line 1468 of file Millard2016TorqueMuscle.h.
◆ mTvCurve
|
private |
Definition at line 1470 of file Millard2016TorqueMuscle.h.
◆ mTvLambda
|
private |
Definition at line 1471 of file Millard2016TorqueMuscle.h.
◆ mTvLambdaMax
|
private |
This is set to the maximum eccentric value of the torque-velocity curve so that the optimization problem of solving for mTaLambda, mTvLambda & mMaxActiveIsometricTorque is convex. This parameter is updated in updateTorqueMuscleCurves
Definition at line 1478 of file Millard2016TorqueMuscle.h.
◆ mUseTabularMaxActiveIsometricTorque
|
private |
Definition at line 1491 of file Millard2016TorqueMuscle.h.
◆ mUseTabularOmegaMax
|
private |
Definition at line 1490 of file Millard2016TorqueMuscle.h.
◆ mUseTabularTorqueVelocityMultiplierAtHalfOmegaMax
|
private |
Definition at line 1492 of file Millard2016TorqueMuscle.h.
The documentation for this class was generated from the following file:
- addons/muscle/Millard2016TorqueMuscle.h